2019.05.17
1. Raman spectro
https://www.nanophoton.net/raman-spectroscopy/lessons/lesson-1
2. Tool
- Learn vocabulary by the Free Dictionary
- Tra từ đồng nghĩa (Synonyms) và trái nghĩa (Antonyms) tại Thesaurus
- Xây dựng dữ liệu NC:
(1) Tìm kiếm và tải báo
tại 1 thời điểm tìm về cập nhật dưới 100 bài báo liên quan đến vấn đề NC
(- Lưu ý vấn đề từ khóa - nguồn thông tin - tạp chí chuyên ngành)
(2) Scan abstract
- đọc abstract và tên = nội dung nghiên cứu > hình dung được bức tranh nghiên cứu hiện tại
- Xây dựng mydatase base dựa trên cơ sở này. (Endnote, Mendeley, Zotero, etc)
- Viết background và State of the art
(3) Cập nhật thụ động bài báo = đăng ký tạp chí chuyên ngành - cập nhật trends nghiên cứu.
- Ví dụ (list tạp chí nghiên cứu các vấn đề)
3. Request topic:
-
SMS
Have a nice day!
Thứ Hai, 20 tháng 5, 2019
Thứ Tư, 15 tháng 5, 2019
PCR amplification not working?
By the way in the PCR reaction include:
DNA template (pure or include inhibition compounds)
Primer
Master mix (include dNTP, DNA polymerase, buffer - it could be optimized)
The reaction happen in thermometer:
Denaturing - when the double-stranded template DNA is heated to separate it into two single strands.
Annealing - when the temperature is lowered to enable the DNA primers to attach to the template DNA.
Extending - when the temperature is raised and the new strand of DNA is made by the Taq polymerase enzyme
notice:
Denaturing depend on enzyme - it could be 94oC eg. Dreamtaq or 98oC eg. Phusion
Annealing - it depend on primer to decide the opt temperature of annealing - low temperature easy to hold primer with DNA but less specific - high temperature need more link between primer and DNA to hold so it more specific. (from it possibly from 48oC to 64oC depend on each primer and ration AT-GC, more GC should be higher temp- was calculating by online tool)
Extending - 72oC for 1-2min depend on speech reaction of enzyme (1kb/min or 1,5kb/min) and the length of product (500bp or 1kbp) to decide the opt time to extend.
- the temperature to optimize- form low to high - to define the high temp give the acurrency result.
- the amount of DNA template (less or more) define easy or difficult to annealing the pcr products (use raw extract DNA in phenol-chloroform or use purified DNA)
- the component of reaction should be control and avoid any inhibition to PCR.
DNA template (pure or include inhibition compounds)
Primer
Master mix (include dNTP, DNA polymerase, buffer - it could be optimized)
The reaction happen in thermometer:
Denaturing - when the double-stranded template DNA is heated to separate it into two single strands.
Annealing - when the temperature is lowered to enable the DNA primers to attach to the template DNA.
Extending - when the temperature is raised and the new strand of DNA is made by the Taq polymerase enzyme
notice:
Denaturing depend on enzyme - it could be 94oC eg. Dreamtaq or 98oC eg. Phusion
Annealing - it depend on primer to decide the opt temperature of annealing - low temperature easy to hold primer with DNA but less specific - high temperature need more link between primer and DNA to hold so it more specific. (from it possibly from 48oC to 64oC depend on each primer and ration AT-GC, more GC should be higher temp- was calculating by online tool)
Extending - 72oC for 1-2min depend on speech reaction of enzyme (1kb/min or 1,5kb/min) and the length of product (500bp or 1kbp) to decide the opt time to extend.
- the temperature to optimize- form low to high - to define the high temp give the acurrency result.
- the amount of DNA template (less or more) define easy or difficult to annealing the pcr products (use raw extract DNA in phenol-chloroform or use purified DNA)
- the component of reaction should be control and avoid any inhibition to PCR.
Primers for amplifying and sequencing fungal rDNA ITS region
PCR primers
To identify ectomycorrhizal (ECM) fungi, ITS1-ITS4, ITS1F-ITS4, and ITS1F-ITS4B are the most widely used primer pairs for PCR. However, using default annealing temperature of 55 degrees centigrade, we have met several difficulties using all of the primer pairs mentioned above.
- ITS1-ITS4 is a pair of universal primers that co-amplifies angiosperm DNA. In case the DNA is extracted from sporocarps, pure cultures, or ECM of conifers, this primer pair amplifies the highest amount of target (ca 600 bp). To our knowledge, ITS1-ITS4 can amplify DNA from nearly all (ECM) fungi.
- ITS1F-ITS4. ITS1F is a fungal specific primer. However, together with ITS4, ITS1F can result in weak amplification of highly concentrated pure angiosperm DNA. We have not observed plant bands on gel (slower) if DNA is extracted from ECM. A problem with ITS1F-ITS4 is gel smearing, that is production of numerous hardly separable bands. Raising annealing temperature a few degrees might resolve this problem. ITS1F tends to result in less target DNA compared to ITS1, but it has proved efficient in amplifying all ECM fungi.
- ITS1F-ITS4B is a pair of strongly basidiomycete-specific primers. In addition to asco-mycetous ECM fungi, Sebacinaceae, Atheliaceae, and some Cortinariaceae species cannot be amplified. We strongly disencourage usage of ITS1F-ITS4B for ECM community studies.
ITS1 or ITS1F together with LR21 appear the most promising primer combination that does not co-amplify plant DNA. These primer pairs amplify nearly all ECM fungi, typically resulting in some 850-900 bp PCR products.
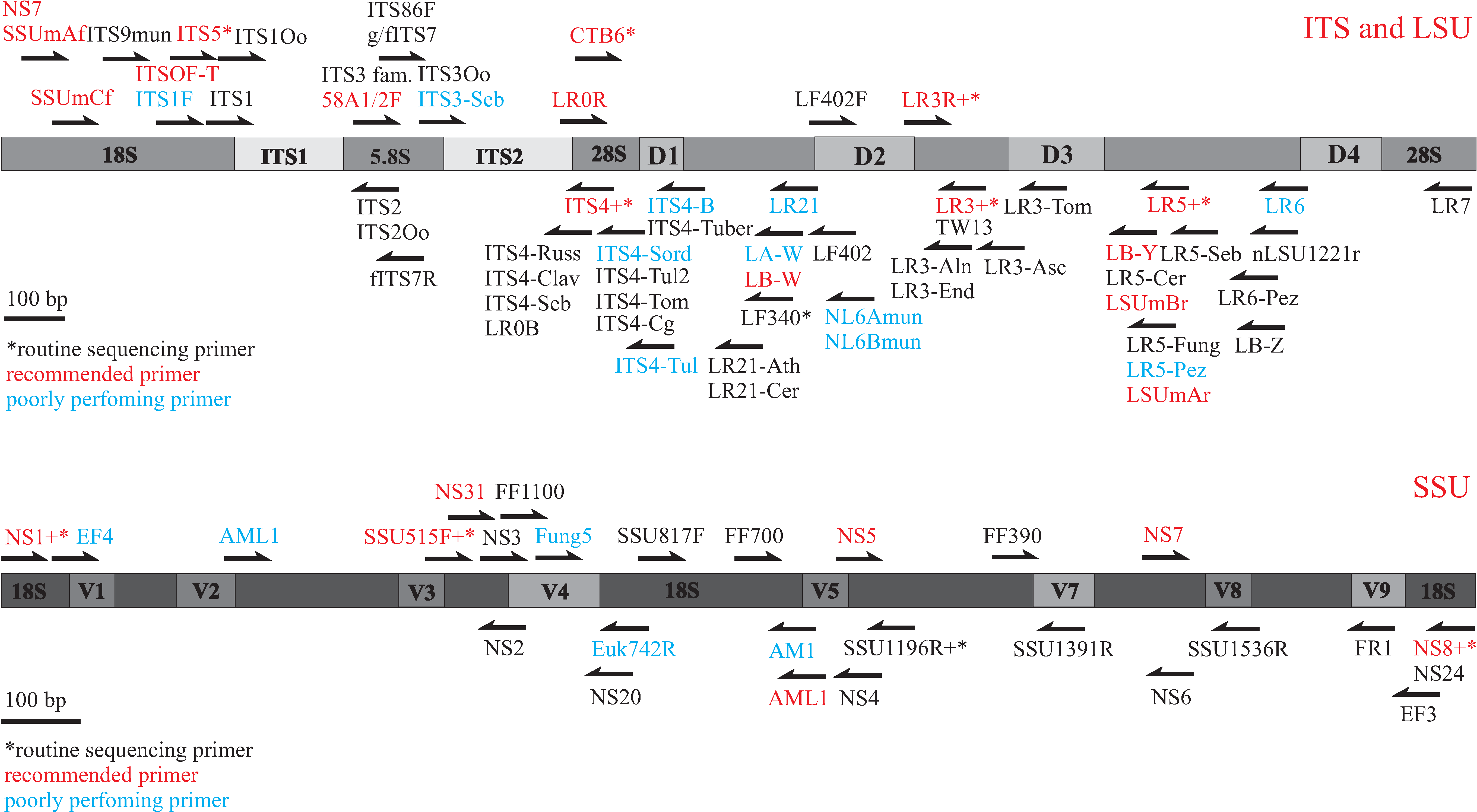
Sequencing primers
In sequencing we have usually applied primers ITS1, ITS2, ITS3, ITS4. ITS2 and ITS3 usually give the best chromatograms. In case of amplification using primer pair ITS1(F)-LR21, ITS3 gives ca 550 bp sequence, covering both ITS2 and ca 200 bp of 28S rDNA. The latter offers more taxonomic opportunities in cases where ITS2 is not matched by other taxa. Sequencing the whole 850 bp with primers ITS1 and LR21 appears to be one of the best solutions.
| Position in gene | Sequence | Gene/locus | Direction | Target | Remarks | Reference | |
| Universal, fungal primers rDNA | |||||||
| 5.8S_Fungi | CAAGAGATCCGTTGTTGAAAGTT | ITS | rev | Fungi | for paleo-DNA | Epp et al. 2012 | |
| 58A1F | 40 | GCATCGATGAAGAACGC | ITS | fwd | Universal | upgrade of ITS3, excludes Glomeromycota | Martin & Rygiewicz 2005 |
| CTB6 | 39 | GCATATCAATAAGCGGAGG | LSU | fwd | Fungi | for sequencing LR0R products | Garbelotto et al. 1997 |
| ITS1 | 30 | TCCGTAGGTGAACCTGCGG | ITS | fwd | Fungi | excludes Sordariomycetes | White et al. 1990 |
| ITS1F | 90 | CTTGGTCATTTAGAGGAAGTAA | ITS | fwd | Fungi | excludes basal fungi and Tulasnella | Gardes & Bruns 1993 |
| ITS1Fngs | 90 | GGTCATTTAGAGGAAGTAA | ITS | fwd | Fungi | excludes basal fungi and Tulasnella | Tedersoo et al. 2015a,b |
| ITS1ngs | 30 | TCCGTAGGTGAACCTGC | ITS | fwd | Fungi | excludes Sordariomycetes | Tedersoo et al. 2015a,b |
| ITS2 | 40 | GCTGCGTTCTTCATCGATGC | ITS | fwd | Universal | excludes some Agaricales, Tremellales | White et al. 1990 |
| ITS3 | 40 | GCATCGATGAAGAACGCAGC | ITS | fwd | Universal | excludes Cantharellales | White et al. 1990 |
| ITS3mix1-5 | 40 | CANCGATGAAGAACGYRG | ITS | fwd | Universal | excludes Tulasnellaceae | Tedersoo et al. 2014 |
| ITS4 | 40 | TCCTCCGCTTATTGATATGC | ITS | rev | Universal | has central mismatches to some groups and Metazoa and Archaeorhizomycetes | White et al. 1990 |
| ITS4ngs | 40 | TCCTSCGCTTATTGATATGC | ITS | rev | Universal | has central mismatches to some groups and Metazoa and Archaeorhizomycetes | White et al. 1990 |
| ITS5 | 50 | GGAAGTAAAAGTCGTAACAAGG | ITS | fwd | Universal | best sequencing primer for ITSOf/1F products | White et al. 1990 |
| fITS7 | 70 | GTGARTCATCGAATCTTTG | ITS | fwd | Fungi | excludes ca 2% minor groups | Ihrmark et al. 2012 |
| fITS7R | 70 | CAAAGATTCGATGAYTCAC | ITS | rev | Fungi | excludes ca 2% minor groups | L. Tedersoo, unpublished |
| gITS7 | 70 | GTGARTCATCGARTCTTTG | ITS | fwd | Fungi | excludes ca 2% minor groups | Ihrmark et al. 2012 |
| ITS86F | 70 | GTGAATCATCGAATCTTTGAA | ITS | fwd | Fungi, Plants | excludes ca 2% minor groups and Tulasnellaceae | Turenne et al. 1998 |
| fITS9 | 55 | GAACACAGCGAAATGTGA | ITS | fwd | Fungi | excludes ca 4% minor groups | Ihrmark et al. 2012 |
| ITS9mun | 190 | TGTACACACCGCCCGTCG | ITS | fwd | Fungi, Plants | little used | Egger 1995 |
| ITSOF | 90 | ACTTGGTCATTTAGAGGAAGT | ITS | fwd | Fungi | outperforms ITS1F | Tedersoo et al. 2008 |
| LF340 | 336 | TACTTGTKCGCTATCGG | ITS | rev | Universal | for sequencing LB-W products | Tedersoo et al. 2008 |
| LF402 | 402 | TTCCCTTTYARCAATTTCAC | ITS/LSU | rev | Fungi | excludes chytrids, Basidio-yeasts,etc. | Tedersoo et al. 2015a,b |
| LF402F | 402 | GTGAAATTGYTRAAAGGGAA | LSU | fwd | Fungi | excludes chytrids, Basidio-yeasts,etc. | Tedersoo et al. 2015a,b |
| LR0B | 15 | GGTAGTCCTACCTGATTTG | ITS | rev | Basidiomycota | excludes several groups incl Sebacinales | L. Tedersoo, unpublished |
| LR0R | 24 | ACCCGCTGAACTTAAGC | LSU | fwd | Universal | widely used for LSU | Hopple & Vilgalys 1994 |
| LR21 | 370 | ACTTCAAGCGTTTCCCTTT | ITS/LSU | rev | Fungi | excludes many Asco- and Basidiomycota | Hopple & Vilgalys 1994 |
| LR3 | 670 | CCGTGTTTCAAGACGGG | ITS/LSU | rev | Universal | widely used for LSU | Hopple & Vilgalys 1994 |
| LR3R | 670 | GTCTTGAAACACGGACC | LSU | fwd | Universal | widely used for LSU | Hopple & Vilgalys 1994 |
| LR5 | 990 | TCCTGAGGGAAACTTCG | LSU | rev | Universal | widely used for LSU | Hopple & Vilgalys 1994 |
| LR5-Fung | 900 | CGATCGATTTGCACGTCAGA | LSU | rev | Fungi | excludes plants, alveolates and rhizarians, but not stramenopiles or animals | Tedersoo et al. 2008 |
| LR6 | 1165 | CGCCAGTTCTGCTTACC | LSU | rev | Universal | fair performance | Hopple & Vilgalys 1994 |
| LR7 | 1475 | TACTACCACCAAGATCT | LSU | rev | Universal | widely used for LSU | Hopple & Vilgalys 1994 |
| nLSU1221r | 1210 | CTAGATGAACYAACACCTT | LSU | rev | Fungi | not enough tested | Schadt et al. 2003 |
| NS7a | 1430 | CAATAACAGGTCTGTGATGC | SSU/ITS | fwd | Universal | widely used | White et al. 1990 (modified) |
| TW13 | 635 | GGTCCGTGTTTCAAGACG | ITS/LSU | rev | Universal | analogue of LR3 | T.J. White, unpublished |
| TW14 | 950 | GCTATCCTGAGGGAAACTTC | LSU | rev | Universal | analogue of LR5 | T.J. White, unpublished |
| Fungal clade-specific primers rDNA | |||||||
| ITS3Seb | 150 | TGAGTGTCATTGTAATCTCAC | ITS | fwd | Sebacinales | excludes ca 20% of target Sebacinales | M.Berbee, unpublished |
| ITS4B | 200 | CAGGAGACTTGTACACGGTCCAG | ITS | rev | Basidiomycota | excludes ca 40% of target taxa | Gardes & Bruns 1993 |
| ITS4B1 | 200 | CAAGRGACTTRTACACGGTCCA | ITS | rev | Basidiomycota | update of ITS4B, still excludes 20% of taxa | Tedersoo et al. 2007 |
| ITS4-Cg | 100 | CACATGGCAARGGCAACCG | ITS | rev | Cenococcum | for improved sequence quality | Bahram et al. 2011 |
| ITS4-Clav | 20 | GGTAGTCCCACCTGATTC | ITS | rev | Clavulina, Cantharellus p.p. | for improved sequence quality | Tedersoo et al. 2011 |
| ITS4-GloeT | 200 | CACAGAAAGCACTTCTCTAA | ITS | rev | Gloeotulasnella | for improved sequence quality | L. Tedersoo, unpublished |
| ITS4-Russ | 20 | AGCGGGTAGTCTCACCC | ITS | rev | Russulaceae | for improved sequence quality | Tedersoo et al. 2011 |
| ITS4-Seb | 20 | TCAGCGGGTARTCCTACTC | ITS | rev | Sebacina clade A | for improved sequence quality | Tedersoo et al. 2011 |
| ITS4-Sord | 100 | CCCGTTCCAGGGAACTC | ITS | rev | Sordariomycetes | for improved sequence quality | Tedersoo et al. 2009 |
| ITS4-Tom | 150 | AACTCGGACGACCAGAGGCA | ITS | rev | Tomentella, Thelephora | excludes 5-10% of Thelephoraceae | Tedersoo et al. 2011 |
| ITS4-Tuber | 230 | CTCGACTCGTAGAAGRCACT | ITS | rev | Tuberaceae | not enough tested | Bonito et al. 2013 |
| ITS4-Tul | 120 | CCGCCAGATTCACACATTGA | ITS | rev | Tulasnellaceae | excludes ca 30% of Tulasnellaceae! | Taylor & McCormick, 2008 |
| ITS4-Tul2 | 50 | TTCTTTTCCTCCGCTGAWTA | ITS | rev | Tulasnella (divergent) | complements to ITS4 and ITS4-Tul for divergent Tulasnellas | Oja et al. 2015 |
| LA-W | 360 | CTTTTCATCTTTCGATCACTC | ITS | rev | Ascomycota | excludes some yeasts, fair PCR product | Tedersoo et al. 2008 |
| LB-W | 360 | CTTTTCATCTTTCCCTCACGG | ITS | rev | Basidiomycota, Zygomycota, Plantae | excludes smuts, some Cantharellus | Tedersoo et al. 2008 |
| LB-wR | 350 | GCGAACAAGTACCGTGAGG | LSU | fwd | Basidiomycota, Zygomycota, Plantae | excludes smuts, some Cantharellus | L. Tedersoo, unpublished |
| LB-y | 870 | TTTGCACGTCAGAATCGCTA | LSU | rev | Basidiomycota (excludes Plantae, other fungi) | good for root tip LSU (excludes some Tulasnella) | Tedersoo et al. 2008 |
| LB-z | 1120 | AAAAATGGCCCACTAGAAACT | LSU | rev | Basidiomycota | good for root tip LSU, excludes some Cantharellus | Tedersoo et al. 2008 |
| LB-Z-Sord | 1020 | GTTTGAGAATGGATGAAGGC | LSU | rev | Sordariomycetes | not enough tested | Tedersoo et al. 2009 |
| LR0R-Tul2 | 40 | CGTTGATTTAAGCATATTAWTC | LSU | fwd | Tulasnella (divergent) | reverse of ITS4-Tul2; not enough tested | L. Tedersoo, unpublished |
| LR21-Ath | 280 | CCAAACAACTCGACTCTTC | ITS | rev | Atheliales | not enough tested | L. Tedersoo, unpublished |
| LR21-Cenoc | 460 | GATGAGCAACATCAGGCAG | ITS | rev | Cenococcum | not enough tested | L. Tedersoo, unpublished |
| LR21-Cer | 300 | CGACTCGTTGAGAGCACAA | ITS | rev | Ceratobasidiaceae | not enough tested | Tedersoo et al. 2011 |
| LR3-Aln | 630 | CCTSAGCACGAACGTGGTA | ITS/LSU | rev | Alnicola, Hebeloma, Entoloma, Inocybe p. parte | for improved sequence quality | L. Tedersoo, unpublished |
| LR3-Asc | 680 | CACYTACTCAAATCCWAGCG | LSU | rev | Ascomycota | not enough tested | Tedersoo et al. 2009 |
| LR3-Cenoc | 580 | TTCAGGCTGGCCGCATTTC | ITS/LSU | rev | Cenococcum | not enough tested | L. Tedersoo, unpublished |
| LR3End | 650 | AYCATTAMGTCAGCGACCTA | LSU | rev | Endogonales | not enough tested | L. Tedersoo, unpublished |
| LR3-Pez | 930 | CMTCRGGATCGGTCGATGG | ITS/LSU | rev | Pezizales | excludes 20% of Pezizales | Tedersoo et al. 2011 |
| LR3-Tul | 570 | GACTCGCATGCAAGGTRCA | ITS/LSU | rev | Tulasnellaceae | excludes 20% of Tulasnellaceae | L. Tedersoo, unpublished |
| LR5-Cer | 860 | CTCTGGCTTCACCCTATG | ITS/LSU | rev | Ceratobasidiaceae | not enough tested | L. Tedersoo, unpublished |
| LR5-Fung | 880 | CGATCGATTTGCACGTCAGA | LSU | rev | Fungi, Metazoa (excl. Plantae) | good for plant tissues | Tedersoo et al. 2008 |
| LR5-Seb | 1000 | ATTCGCTTTACCGCACAAGG | LSU | rev | Sebacinales | not enough tested | Tedersoo et al. 2008 |
| LR5-Tom | 740 | CTACCGTAGAACCGTCTCC | ITS/LSU | rev | Tomentella, Thelephora | excludes 10% of Thelephoraceae | Tedersoo et al. 2008 |
| LR6 Leot-Sord | 1100 | AAAATGGCCCACTAGTGTTG | LSU | rev | Leotiomycetes, SordarioM | not enough tested | Tedersoo et al. 2011 |
| LR6-Asc | 1100 | AAAATGGCCCACTAGTAACG | LSU | rev | Ascomycota, v.a. Leot,SordM | not enough tested | Tedersoo et al. 2011 |
| LR6-Pez | 1100 | CCTCATAAAACRAKATCGTTAC | LSU | rev | Pezizales | not enough tested | L. Tedersoo, unpublished |
| LSUmAr | 930 | [composite] | ITS/LSU | rev | Glomeromycota | good for AMF | Krüger et al. 2009 |
| LSUmBr | 840 | [composite] | ITS/LSU | rev | Glomeromycota | good for AMF | Krüger et al. 2009 |
| NL6Amun | 420 | CAAGTGCTTCCCTTTCAACA | ITS | rev | Ascomycota | not specific, excludes many groups; 1-2 mismatches to Archaeorhizomycetes | Egger 1995 |
| NL6Bmun | 420 | CAAGCGTTTCCCTTTCAACA | ITS | rev | Basidiomycota | not specific, excludes many groups | |
| SSUmAf | 300 | [composite] | ITS | fwd | Glomeromycota | good for AMF | Krüger et al. 2009 |
| SSUmCf | 250 | [composite] | ITS | fwd | Glomeromycota | good for AMF | Krüger et al. 2009 |
| Universal, fungal SSU | |||||||
| FF1100 | 580 | CCAGCTCCAATAGCGTATATTA | SSU | fwd | Fungi | Vainio & Hantola 2000 | |
| FF700 | 980 | GATACCGTIGTAGTCT | SSU | fwd | Fungi | Vainio & Hantola 2000 | |
| FF390 | 1290 | CGATAACGAACGAGACCT | SSU | fwd | Fungi | Vainio & Hantola 2000 | |
| FR1 | 1680 | AICCATTCAATCGGTAIT | SSU | rev | Fungi | Vainio & Hantola 2000 | |
| SSU515f | 500 | GTGCCAGCMGCCGCGGTAA | SSU | fwd | Euakryote/Prokaryote | Cross-domain primer | Turner et al. 1999 |
| SSU515Fngs | 500 | GCCAGCAACCGCGGTAA | SSU | fwd | Euakryote/Prokaryote | Tedersoo et al. 2015a,b | |
| SSU0817f | 790 | TTAGCATGGAATAATRRAATAGGA | SSU | fwd | Eukaryote | Borneman & Hartin 2000 | |
| SSU1196R | 1170 | TCTGGACCTGGTGAGTTTCC | SSU | rev | Eukaryote | Borneman & Hartin 2000 | |
| SSU1196Rngs | 1170 | TCTGGACCTGGTGAGTTT | SSU | rev | Eukaryote | Tedersoo et al. 2015a,b | |
| SSU1391R | 1360 | GACGGGCGGTGWGTRCA | SSU | rev | Eukaryote | Turner et al. 1999 | |
| SSU1536R | 1490 | ATTGCAATGCYCTATCCCCA | SSU | rev | Eukaryote | Borneman & Hartin 2000 | |
| EF4 | 120 | GGAAGGGRTGTATTTATTAG | SSU | fwd | Eukaryote | Smit et al. 1999 | |
| EF3 | 1700 | TCCTCTAAATGACCAAGTTTG | SSU | rev | Eukaryote | Smit et al. 1999 | |
| Fung5 | 680 | GTAAAAGTCCTGGTTCCCC | SSU | rev | Fungi | excludes 10% groups incl. ArchaeorhizoM | Smit et al. 1999 |
| AM1 | 1150 | GTTTCCCGTAAGGCGCCGAA | SSU | rev | Glomeromycota | excludes early diverging groups | Helgason et al. 1998 |
| AML1 | 230 | ATCAACTTTCGATGGTAGGATAGA | SSU | rev | Glomeromycota | Lee et al. 2008 | |
| AML2 | 1170 | GAACCCAAACACTTTGGTTTCC | SSU | rev | Glomeromycota | Lee et al. 2008 | |
| NS1 | 30 | GTAGTCATATGCTTGTCTC | SSU | fwd | Eukaryote | White et al. 1990 | |
| NS2 | 560 | GGCTGCTGGCACCAGACTTGC | SSU | rev | Eukaryote | White et al. 1990 | |
| NS7 | 1400 | GAGGCAATAACAGGTCTGTGATGC | SSU | fwd | Eukaryote | White et al. 1990 | |
| NS6 | 1400 | GCATCACAGACCTGTTATTGCCTC | SSU | rev | Eukaryote | White et al. 1990 | |
| NS3 | 560 | GCAAGTCTGGTGCCAGCAGCC | SSU | fwd | Eukaryote | White et al. 1990 | |
| NS4 | 1120 | CTTCCGTCAATTCCTTTAAG | SSU | rev | Eukaryote | White et al. 1990 | |
| NS5 | 1140 | AACTTAAAGGAATTGACGGAAG | SSU | fwd | Eukaryote | White et al. 1990 | |
| NS8 | 1770 | TCCGCAGGTTCACCTACGGA | SSU | rev | Eukaryote | White et al. 1990 | |
| NS20 | 860 | CGTCCCTATTAATCATTACG | SSU | rev | Eukaryote | White et al. 1990 | |
| NS24 | 1760 | AAACCTTGTTACGACTTTTA | SSU | rev | Eukaryote | White et al. 1990 | |
| NS31 | 535 | TTGGAGGGCAAGTCTGGTGCC | SSU | fwd | Fungi | widely used | Simon et al. 1992 |
| Euk742R | 890 | AAATCCAAGAATTTCACCTCT | SSU | rev | Eukaryote | poor performance | Tedersoo et al. 2015a,b |
| Plant ITS primers | |||||||
| RydinF | 70 | GAACCTTATCRTTTAGAGGAAGG | ITS | fwd | Plantae | ||
| Rydin R | 50 | CCGCCAGATTTTCACGCTGGGC | ITS | rev | Plantae | ||
| ITS4Ang | 20 | GTARTCCCGCCTGACCTG | ITS | rev | Angiospermae | ||
| ITS4PL | 260 | TTCCCAAACAACCCGACTCG | ITS | rev | Plantae | matches many fungi | |
| ITS-OP | 70 | ttaTCATTTAGAGGAAGgAg | ITS | fwd | Plantae | plant analogue of ITS1F | |
| PN16 | 400 | TCCCTTTCAACAATTTCACG | ITS/LSU | rev | Plantae | excludes many groups | |
| 18S-IGS | 20 | ACTACTGGCAGGATCAACCAG | IGS | rev | Plantae, Fungi | ||
| Primers for chloroplast markers | |||||||
| trnL-c | . | CGAAATCGGTAGACGCTACG | trnL | fwd | Plantae | ||
| trnL-d | . | GGGGATAGAGGGACTTGAAC | trnL | rev | Plantae | ||
| trn F | . | ATTTGAACTGGTGACACGAG | trnL | rev | Plantae | ||
| trn E | . | GGTTCAAGTCCCTCTATCCC | trnL | fwd | Plantae | ||
| trn H | . | CGCGCATGGTGGATTCACAATCC | trnL | fwd | Plantae | ||
| psbA | . | GTTATGCATGAACGTAATGCTC | trnL | rev | Plantae | ||
| atpF | . | GAAGTAGTAGGATTGATTCTC | rbcL | rev | Plantae | ||
| rbcL | . | CCCTACAACTCATGAATTAAG | rbcL | fwd | Plantae | ||
| Oomycete primers | |||||||
| ITS1Oo | 0 | GGAAGGATCATTACCACAC | ITS | fwd | Oomycota (99%) | L. Tedersoo, unpublished | |
| ITS2Oo | 40 | GCAGCGKTCTTCATCGRTGT | ITS | fwd | Oomycota (99%) | L. Tedersoo, unpublished | |
| ITS3Oo | 150 | AGTATGYYTGTATCAGTGTC | ITS | fwd | Oomycota (99%) | T. Riit, unpublished | |
| ITS3Perofascia | 140 | CCACCTATGCTACGCTATG | ITS | fwd | T. Riit, unpublished | ||
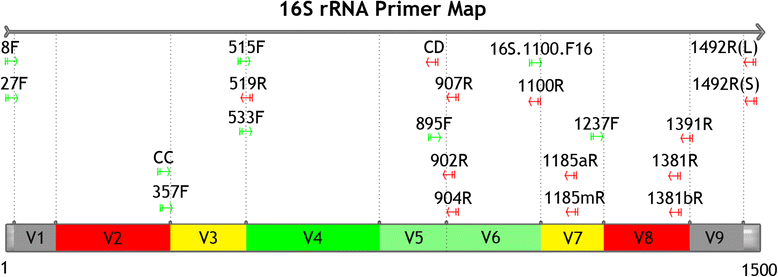
Primer for bacterial

Angela M Orshinsky added an answer
For fungi: http://onlinelibrary.wiley.com/doi/10.1111/j.1755-0998.2009.02635.x/full They describe and evaluate the benefits of different standard primer targets and make a good case for ITS4 and ITS5 as universal fungal primers.
The authors prefer chaperonin as a target for bacterial species identification from environmental samples.

Đăng ký:
Bài đăng (Atom)